Task Description
Clinical narratives are abundant with named entity mentions for clinical conditions, anatomical sites, medications, and procedures. Many surface forms are representations of the same concept. In biomedicine, there are rich lexical and ontological resources that can be leveraged when building applications. The Unified Medical Language System (UMLS) represents over 130 lexicons/thesauri with terms from a variety of languages. The UMLS Metathesaurus integrates resources such as SNOMED-CT, ICD9, and RxNORM that are used worldwide in clinical care, public health, and epidemiology. In addition, the UMLS also provides a semantic network in which every concept in the Metathesaurus is represented by its Concept Unique Identifier (CUI) and is semantically typed (Bodenreider and McCray, 2003). In addition to recognizing and normalizing named entities in clinical text, one fundamental task of clinical natural language processing is to identify the potential modifiers attached to specific named entities, such as negation and uncertainty.
The Analysis of Clinical Task is split into two tasks, one on named entity recognition, and one on template slot filling for the named entities. Participants can submit to either or both tasks.
Task 1: Disorder Identification
In the disorder identification task, the goal is to recognize the span of a disorder mention and its normalization to a unique CUI in the UMLS/SNOMED-CT terminology in a set of clinical notes. (UMLS/SNOMED-CT is the set of CUIs in UMLS restricted to concepts that are part of the SNOMED-CT vocabulary).
Here are a few examples—more are provided in the annotation guidelines and in the page on Datasets. Given the following three sentences:
- The rhythm appears to be atrial fibrillation.
- The left atrium is moderately dilated.
- 53 year old man s/p fall from ladder.
The spans of the disorder mentions are identified as follows: In examples 1. and 3., the phrases atrial fibrillation and fall from ladder fall in the disorder semantic group in the UMLS. Example 2. is a case of discontigous mentions represented by left atrium...dialated. This phenomena where a discontiguous phrase is the best representative of the disorder occurs more commonly in the clinical domain than in the general domain, and therefore is annotated as such.
The disorder entities identified in the examples above map to the following CUIs:
- atrial fibrillation - C0004238; UMLS preferred term atrial fibrillation
- left atrium...dilated - C0344720; UMLS preferred term left atrial dilatation
- fall from ladder - C0337212; UMLS preferred term is accidental fall from ladder
Example 1. represents the easiest cases; Example 2. represents instances of disorders as listed in the UMLS are best mapped using disjoint mentions; Example 3. is harder as one has to infer that the description is a synonym of the UMLS preferred term.
In some cases, a disorder mention is present, but there is no good equivalent CUI in UMLS/SNOMED-CT. The disorder is then normalized to “CUI-less”.
Participants are free to use any UMLS resources as well as other supplemental content such as WordNet, Wikipedia, etc. In addition to this, we have made the rest of the MIMIC corpus of clinical notes (from which these notes were sampled for annotation) available as a larger corpus for exploring semi-supervised and unsupervised methods.
Evaluation Metric
Evaluation will be carried out according to the following F-scores:
Strict F-score: a predicted mention is considered a true positive if (i) its predicted span is exactly the same as for the gold-standard mention; and (ii) the predicted CUI is correct. The predicted disorder is considered a false positive if the span is incorrect or the CUI is incorrect.
Relaxed F-score: a predicted mention is a true positive if (i) there is any word overlap between the predicted mention span and the gold-standard span (both in the case of contiguous and discontiguous spans); and (ii) the predicted CUI is correct. The predicted mention is a false positive if the span shares no words with the gold-standard span or the CUI is incorrect.
Task 2: Disorder Slot Filling
For a given disorder mention, there are several attributes one can identify. This task focuses on identifying the normalized value for nine modifiers in a disorder mentioned in a clinical note: the CUI of the disorder (very much like in Task 1), as well as the potential attributes (negation indicator, subject, uncertainty indicator, course, severity, conditional, generic indicator, and body location) as described in Table 1. The Clinical Element Models are the original source of all of the attributes.
Table 1. Disease/Disorder Attribute Types with definitions and norm and cue slot values.
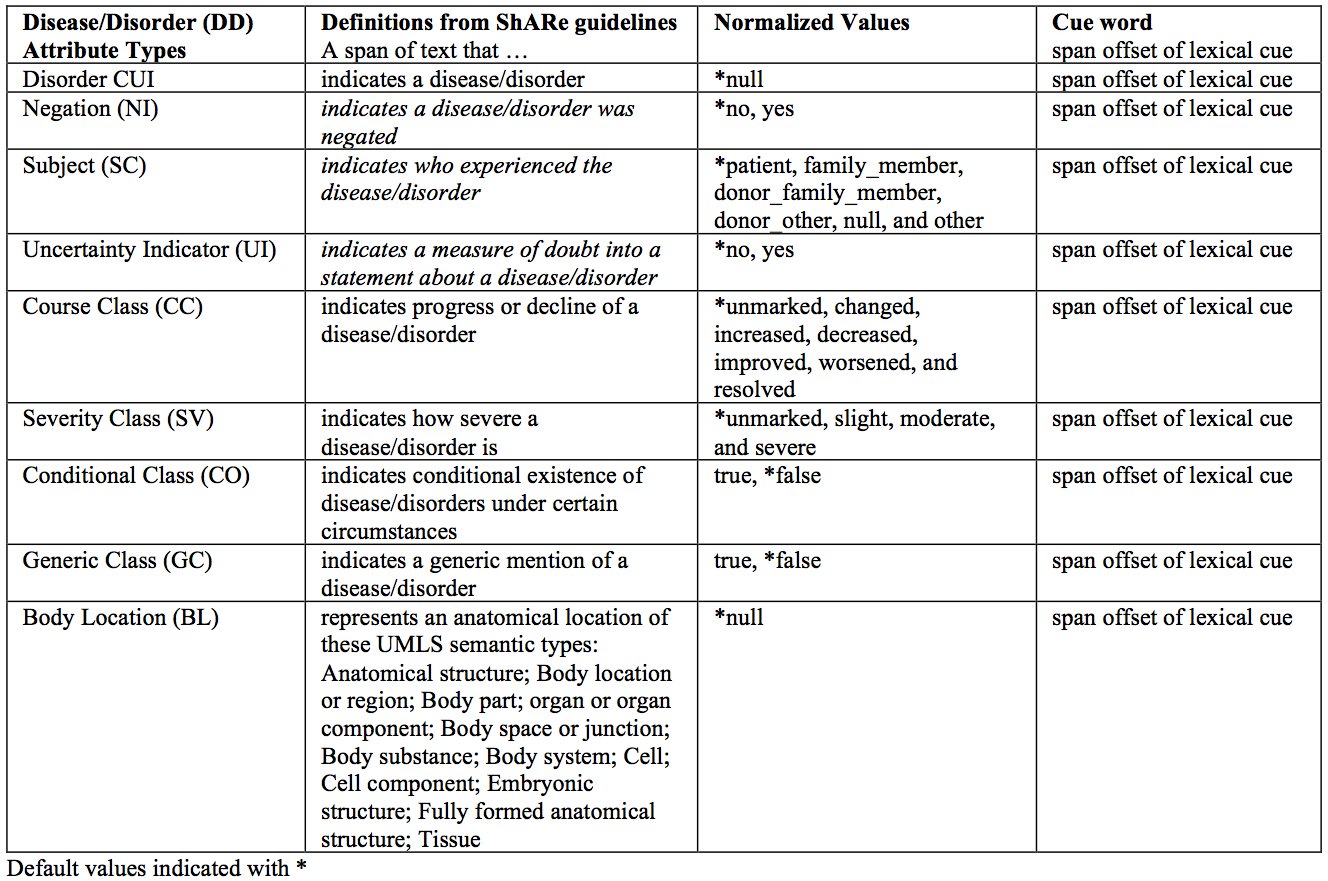
More specifically, the slot-filling task is as follows: given the span (gold-standard or system identified) of a disorder in a clinical note, identify the value for nine slots (CUI of the disorder, negation indicator, subject, uncertainty indicator, course, severity, conditional, generic indicator, and body location) according to their normalized values. Note that there are two aspects to slot filling: cue and normalized value. In this task, we focus on normalized value and ignore cue detection.
There are two subtasks. In both cases, given a disorder span, participants are asked to identify nine attributes related to the disorder. In Task 2a, the gold-standard span(s) of disorders are given. In Task 2b, no gold-standard information is provided; the participants must recognize spans for disorder mentions and fill out the value of the nine attributes.
The training dataset has a pipe-delimited format as follows. The predicted disorders and slots must follow the same format. Since the span fields are not required for the predictions, participants can output a “NULL” token for them.
DD_DocName|DD_Spans|DD_CUI|Norm_NI|Cue_NI|Norm_SC|Cue_SC|Norm_UI|Cue_UI| Norm_CC|Cue_CC|Norm_SC|Cue_SC|Norm_CO|Cue_CO|Norm_GC|Cue_GC|Norm_BL|Cue_BL
Example values for the Normalization slots are shown in Table 2.
Table 2. Attribute types with example sentences and their norm and cue slot values.
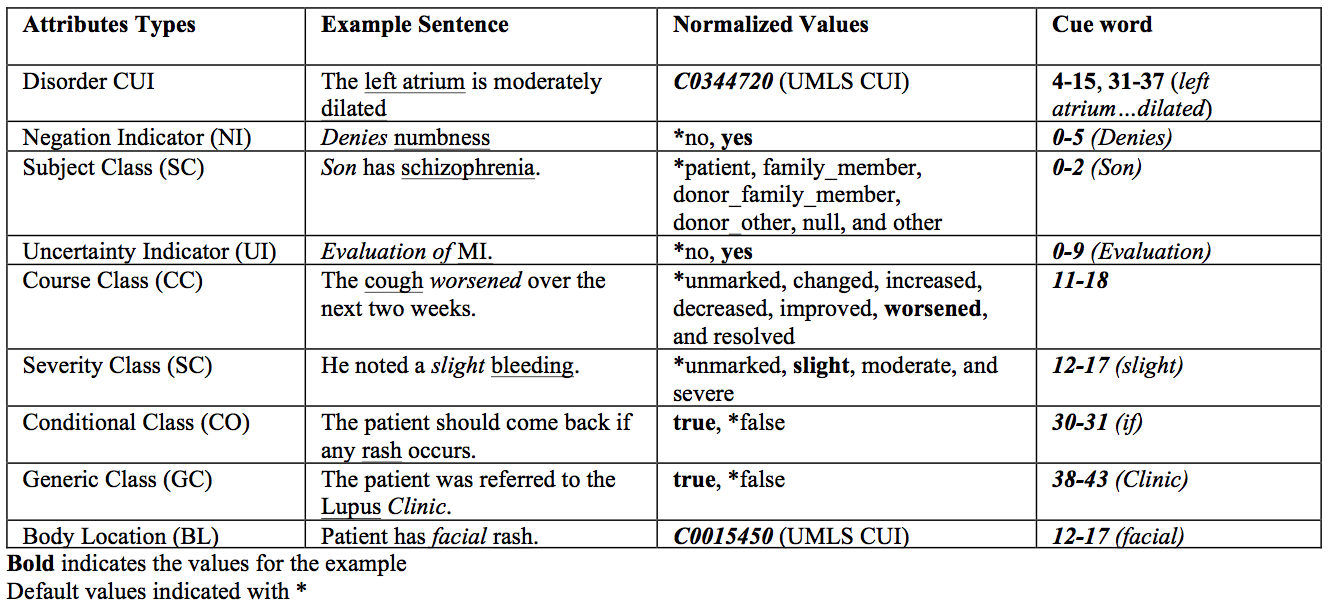
Task 2a – Slot Filling Given Gold-Standard Disorder Spans
In this subtask, the spans of gold-standard disorders are provided to the participants, and the task consists of identifying the values for the nine attributes.
Evaluation Metrics:
The following metrics will be computed (more details on the metrics is available in this document)
Unweighted Accuracy: For each disorder a per-disorder, unweighted accuracy is computed, which represents the ability to identify all the slots for that disorder. The unweighted accuracy is the average of the per-disorder unweighted accuracy over all the disorders in the test set.
Weighted Accuracy: For each disorder, a weighted per-disorder accuracy is computed, which represents the ability to identify all the slots for that disorder. Each gold-standard slot value is pre-assigned a weight based on its prevalence in the training set. The weighted accuracy is the average of the per-disorder weighted accuracy over all the disorders in the test set.
In addition, for information purposes, the following metrics will be computed.
Per-slot Weighted Accuracy: For each slot, a per-slot weighted accuracy is computed, that represents the ability of a system to identify the different values of that slot across all disorders in the test set. These metrics are useful to assess the difficulty of filling a particular slot.
Example: For the following sentence, “The patient has an extensive thyroid history.”, participants are provided the following disease/disorder template with defaults:
00000-000000-DISCHARGE_SUMMARY.txt|30-36|C0040128|no|NULL|patient|NULL|no|NULL| unmarked|NULL| unmarked|NULL|false|NULL|false|NULL|NULL|NULL
Participants will keep or update the Normalization values. For the example sentence, in Task 2a the above template will be modified to:
00000-000000-DISCHARGE_SUMMARY.txt|30-36|C0040128|no|NULL|patient|NULL|no|NULL| unmarked|NULL| unmarked|NULL|severe|NULL|false|NULL|C0040132|29-35
Task 2b – End-to-End System: Disorder Span and Slot Filling
In this subtask, the participants must identify span disorders and their corresponding nine slots. As such the evaluation metrics will take into account both the ability to recognize disorder mentions and their slots.
Evaluation Metrics:
The following metrics will be computed (more details on the metrics is available in this document)
F score for span identification (based on a relaxed definition of the span)
Unweighted Accuracy: The same as the unweighted accuracy described in Task 2a, computed over the true-positive identified disorders
Weighted Accuracy: The same as the weighted accuracy described in Task 2b, computed over the true-positive identified disorders.
In addition, for information purposes, the following metrics will be computed.
Per-slot Weighted Accuracy: For each slot, a per-slot weighted accuracy is computed, that represents the ability of a system to identify the different values of that slot across all disorders in the test set. These metrics are useful to assess the difficulty of filling a particular slot.
Reference:
Bodenreider, O. and McCray, A. 2003. Exploring semantic groups through visual approaches. Journal of Biomedical Informatics, 36(2203): pp. 414-432.
Pradhan, S., Elhadad, N., South, B., Martinez, D., Christensen, L., Vogel, A., Suominen, H., Chapman, W., Savova, G., Evaluating the state of the art in disorder recognition and normalization of the clinical narrative, J Am Med Inform Assoc doi:10.1136/amiajnl-2013-002544